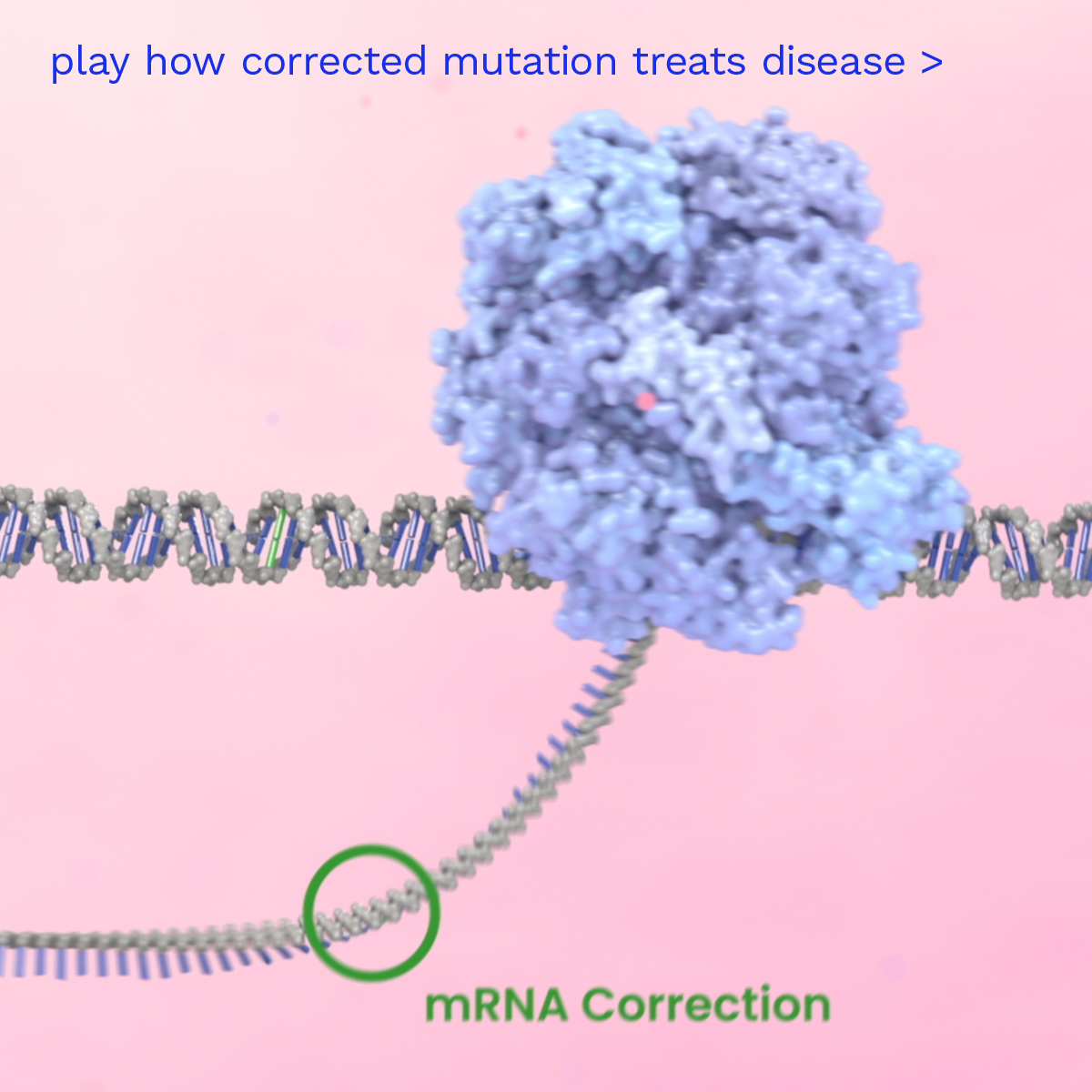
about
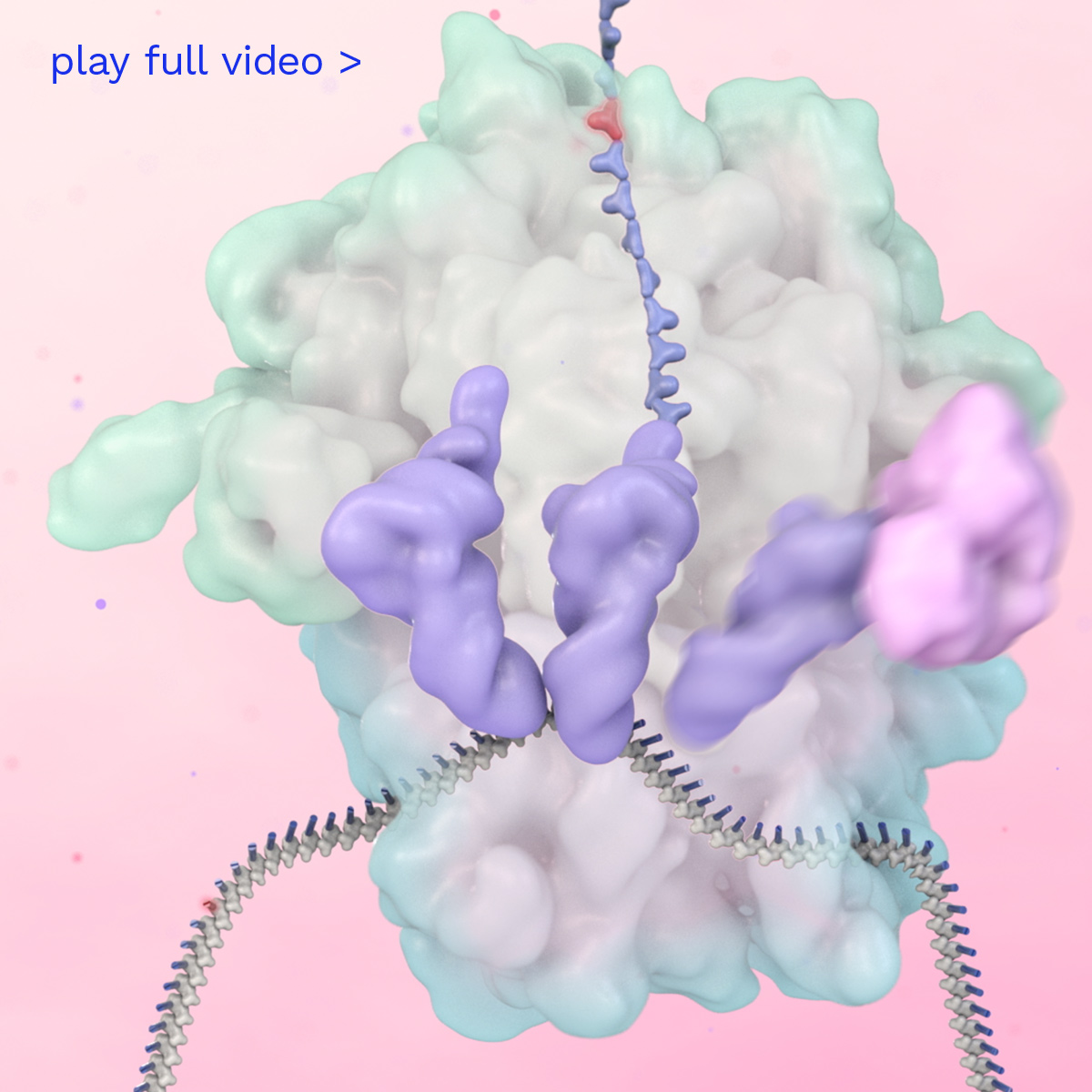
Prime Editing is a next-generation gene editing technology that acts like a DNA word processor, with the power to search and replace genetic sequences at their exact location in the genome, all without making double-strand breaks in DNA.
Prime Editors are designed to produce precise edits and have editing activity across many different tissues, organs and types of cells. Prime Editors work broadly across almost all types of genetic mutations at the natural genomic location, while minimizing unwanted DNA modifications. The Prime Editing platform is designed to be able to produce a wide variety of precise, predictable and efficient genetic outcomes at the targeted pathogenic gene sequence, which we believe will dramatically increase the impact of gene editing for a broad range of therapeutic applications.
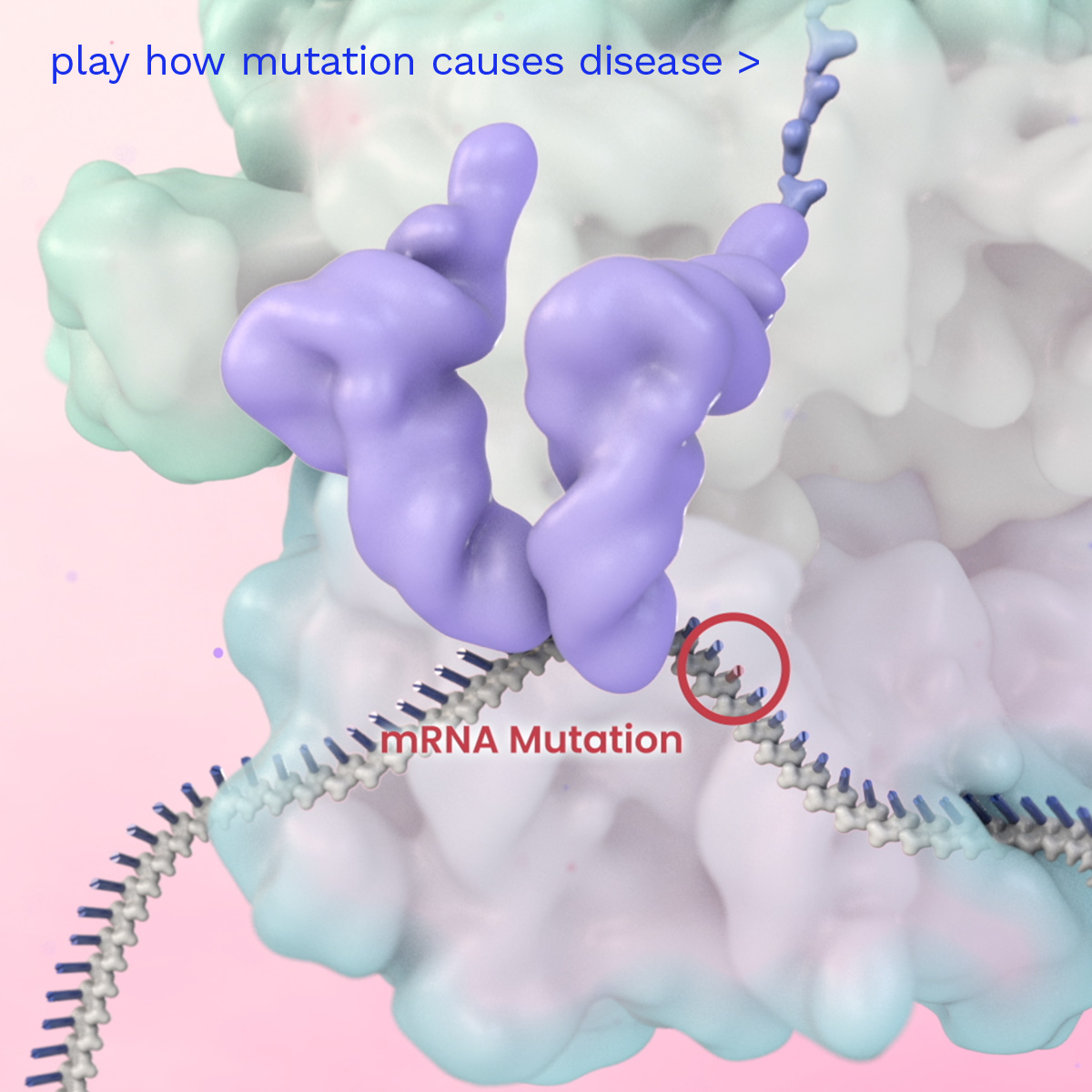
Prime Medicine aims to deliver a new class of differentiated one-time curative genetic therapies to patients to address the widest spectrum of diseases. Watch how it is intended to work inside the human body to address an example of a disease caused by a single base mutation in a gene.
science
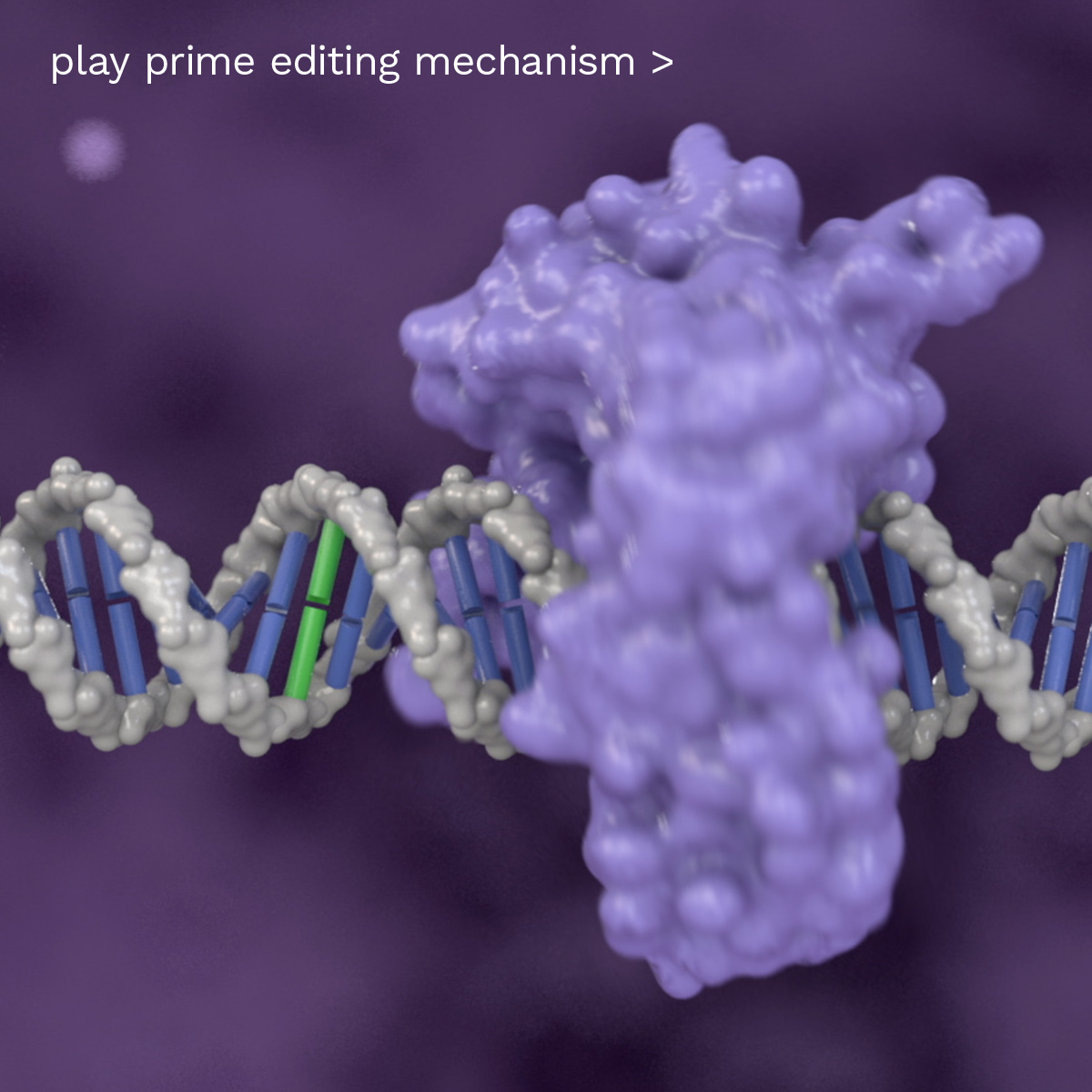
How Prime Editing Works
Programmable for both search and replace





Exemplary Prime Editing process using Cas and RT. Abbreviations: pegRNA = Prime Editing guide RNA; RT = reverse transcriptase; Cas = CRISPR associated protein
Key Features
- Programmable and highly flexible
- High fidelity and specificity
- High editing efficiency
- Minimal off-target activity
- Potential for improved gene function through in situ editing
- Edits in multiple clinically relevant dividing and non-dividing cell-types and organs
- No DNA double-strand breaks
- Validated in multiple independent laboratories worldwide
versatile
Prime Editing can correct all different types of mutations
Point Mutation
Convert any base pair into any other

Point Mutation
Mutated base pairs can be substituted

Insertion Mutation
Extra base pairs can be removed

Deletion Mutation
Missing base pairs can be inserted

Versatile
Prime Editing can make diverse sequence edits at nearly any desired location in the human genome, enabling multiple therapeutic applications. Prime Editors are able to change any base pair to any other base pair to correct all twelve types of single base pair point mutations, delete DNA sequences to correct insertion mutations, and insert DNA sequences to correct deletion mutations. Prime Editors also have the ability to correct genetic mutation hotspots and modify or silence gene expression by altering the regulatory regions of genes, inserting or creating premature stop codons, or modifying splicing sequences. Prime Editors additionally have the capacity to install genetic sequences to protect from disease or to correct genetic variants that increase disease risk.
precise
One prime editing pegRNA can precisely correct the individual mutations found across many patients
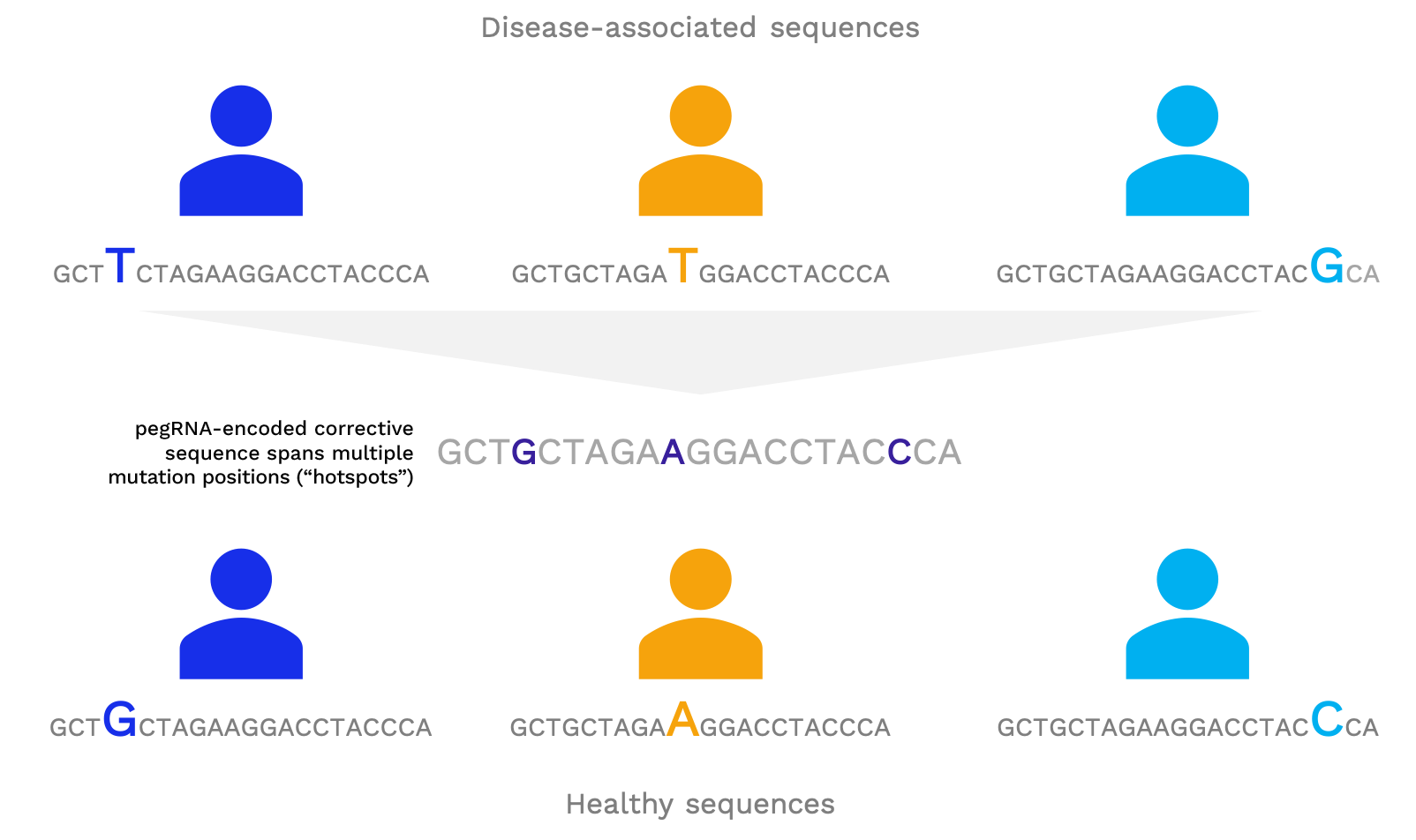
Abbreviations: pegRNA = Prime Editing guide RNA.
Precise
Prime Editing is designed to make only the right edit at the right position within a gene, which greatly minimizes on-target by-products at the site of editing, and results in low or minimal off-target editing in other places in the genome. Our Prime Editors do not create double-strand DNA breaks, which supports the precision of our technology. Prime Editing requires three “edit checks” or places where there must be a match between the editor and the target DNA in order to complete an edit, which we also believe leads to highly specific and precise edits.
dual flap
Dual Flap – an advance on conventional Prime Editing technology
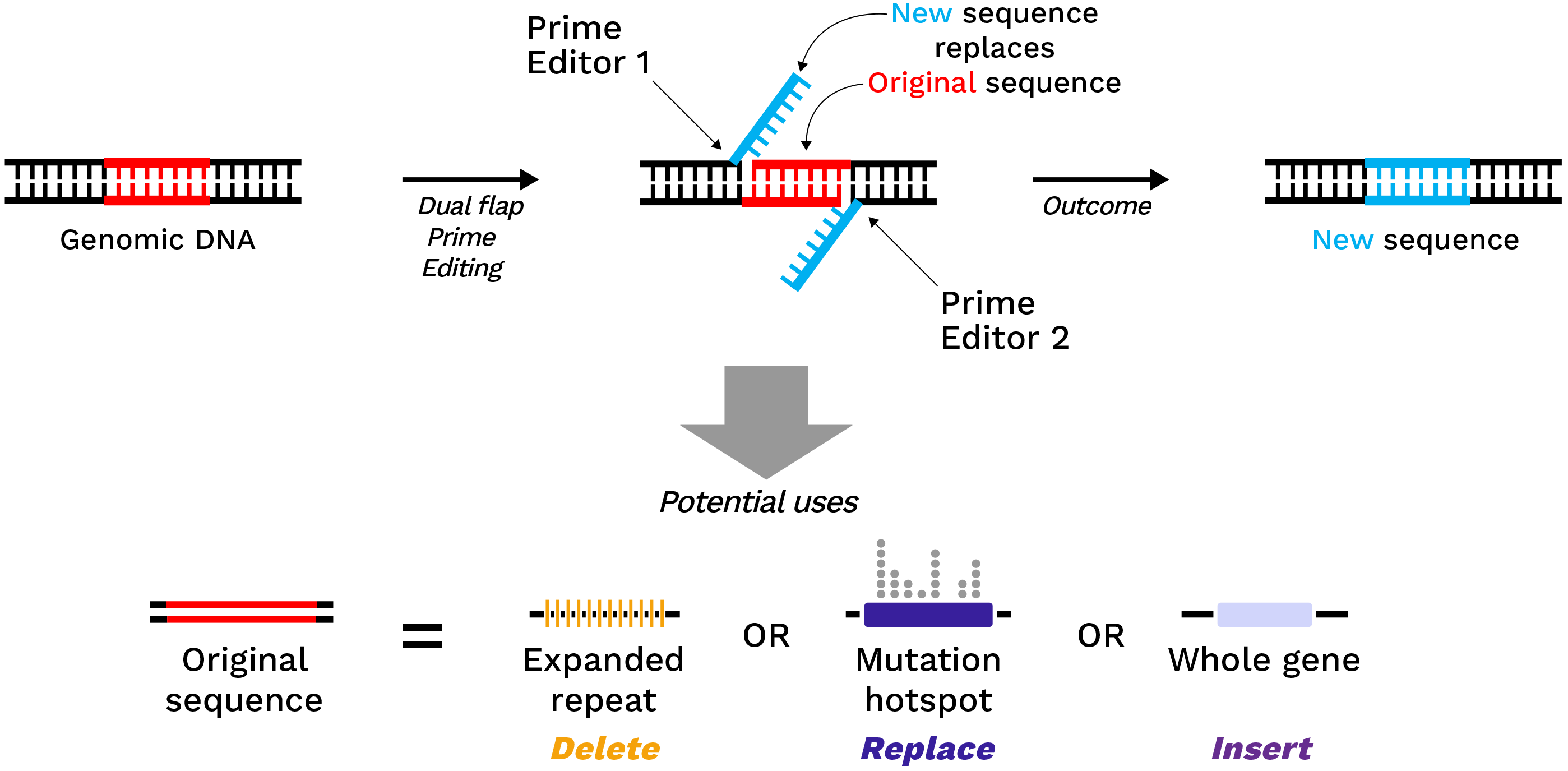
Dual Flap Prime Editing
Dual Flap Prime Editing, also known as Twin Prime Editing, uses two Prime Editors working together to make a broader range of edits at a precise location in the genome. The broader range of edits possible with this advance include looping out and deleting large genomic regions such as those occurring in repeat expansion diseases, correcting large hotspot regions in a gene, replacing or removing small exons, or inserting DNA sequences. All of these edits occur without double-strand breaks, and require the same “edit checks” as in conventional Prime Editing.
efficient
Prime editing corrections occur at the natural place in the genome and are permanent.

Efficient
With a single treatment, Prime Editing could permanently correct disease-causing mutations, resulting in restoration of the gene back to its wild-type, healthy sequence. A corrected gene would persist in an edited cell, working naturally and being passed along to daughter cells, leading to a potentially durable cure or therapeutic outcome. These benefits have the potential for optimal gene regulation, which we believe could result in long-lasting benefits to patients.
passige
PASSIGE™ – further extending Prime Editing to insert gene sized sequences precisely in the genome
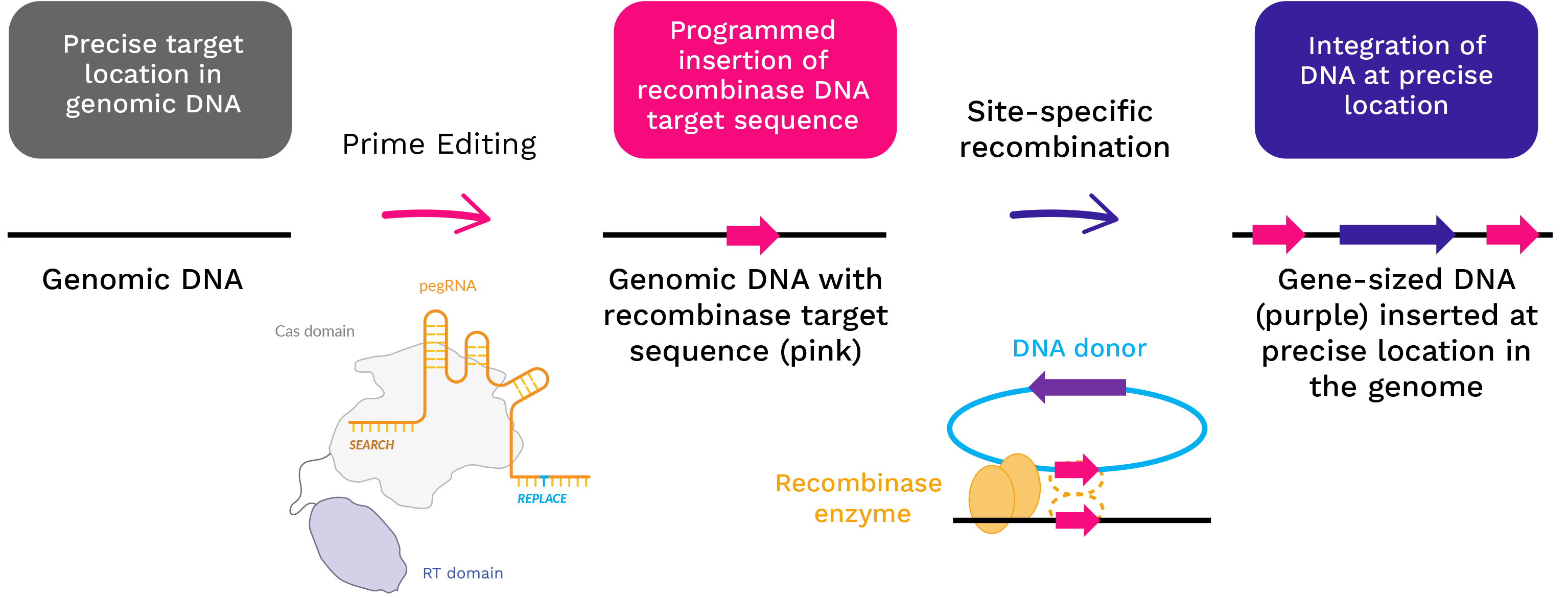
Exemplary Prime Editing process using Cas and RT. Abbreviations: pegRNA = Prime Editing guide RNA; RT = reverse transcriptase; Cas = CRISPR associated protein
PASSIGE – Precise introduction of gene-sized pieces of DNA
By combining Prime Editing with an integrase or site-specific recombinase enzyme we harness the precision of Prime Editing with the ability to introduce large gene-sized cargo into the genome as a potential one-time therapy for patients. This proprietary therapeutic approach, known as Prime Assisted Site Specific Integrase Gene Editing or PASSIGE, further increases the versatility of Prime Editing. We believe PASSIGE broadens the range of permanent genomic edits that Prime Editing can make to treat disease, including the ability to insert entire genes precisely into a patient’s genome.
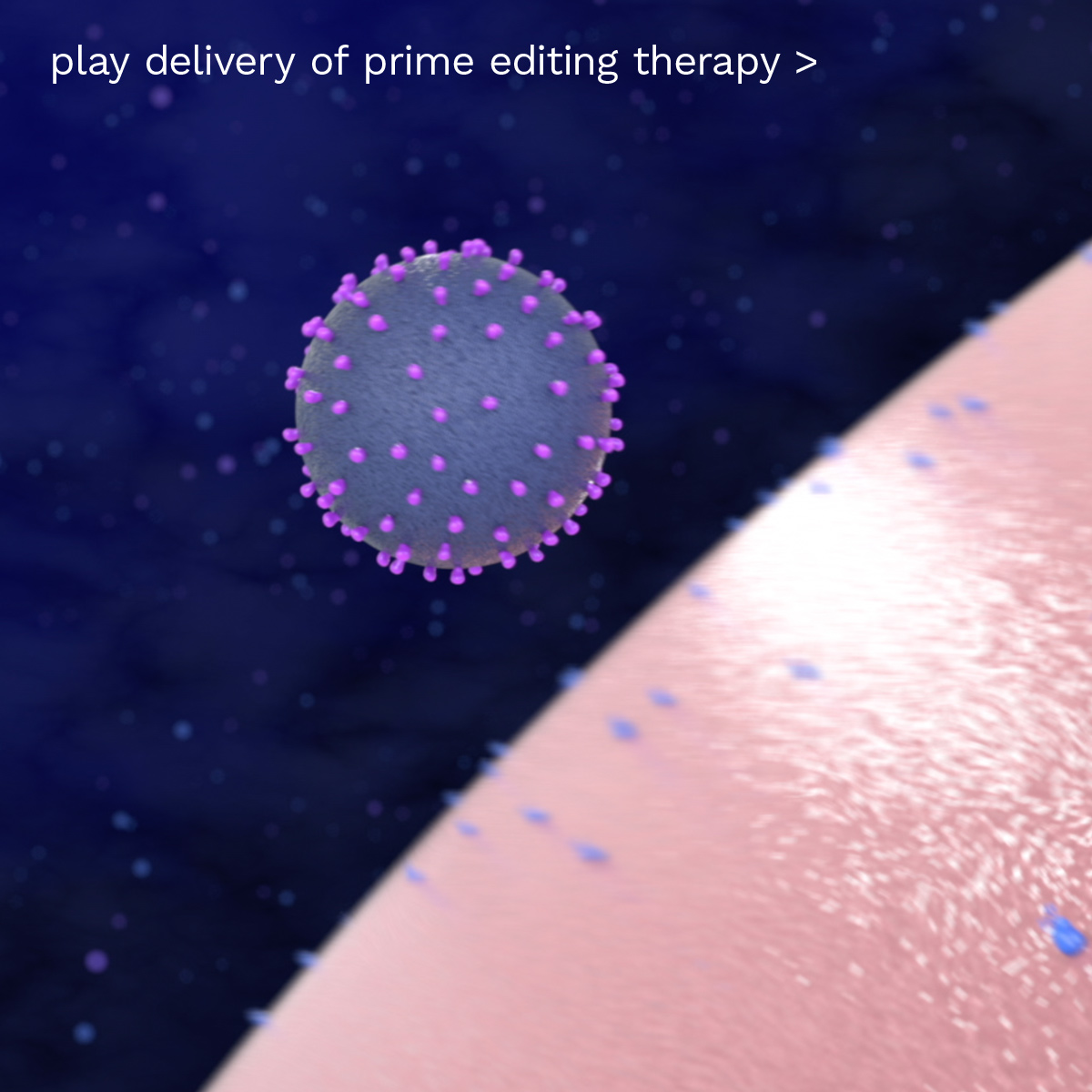
broad
Prime Editing can be delivered to the right place in the body using multiple modalities
Non-viral nanoparticle

RNA-protein complex

Viral vector

Prime-Edited cell therapy

Broad
Prime Editing is a compelling approach for a wide range of therapeutic applications at the genomic level, and can make precise, targeted edits in an array of cell types, tissues and organs. We believe this breadth in applications and ability to target multiple cell types, as well as technological advances such as dual flap and PASSIGE, will enable Prime Editing to bring potentially curative gene editing approaches to a broader set of diseases, beyond rare genetic disease, to address severe, chronic, and acute common diseases with high unmet need, or replace current therapies for disease with a one-time genetic therapy.